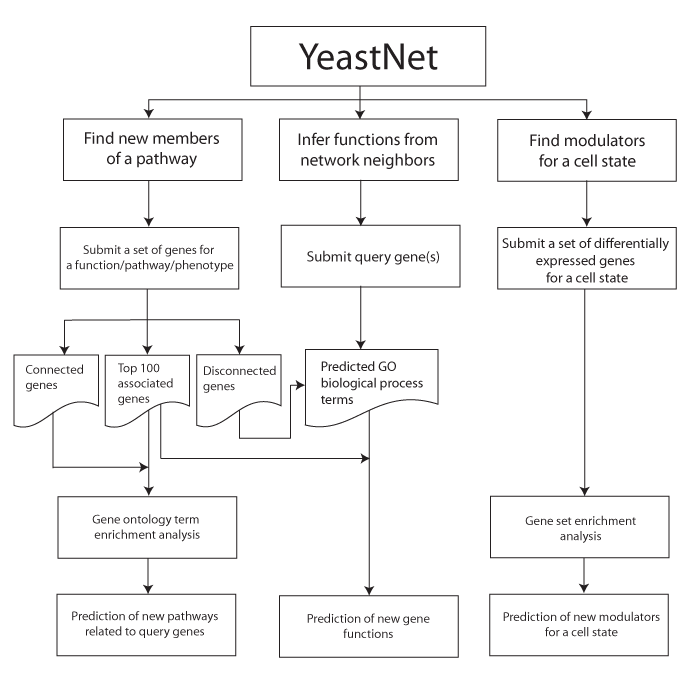
YeastNet provides three options of network search: (i) Find new members of a pathway, (ii) Infer functions from network neighbors, (iii) Find modulators for a cell state.
If you want to identify new candidate genes for a function/pathway/phenotype, you can start search by submitting a set of known genes for the function/pathway/phenotype. These submitted genes are used as 'seed genes' for network search. This search page collects closely connected genes to the seed genes on YeastNet by 'guilt-by-association' method and list the prioritized genes by total edge weight score (sum of log likelihood score) to the seed gene set. Highly ranked genes are good candidates for new members of a function/pathway/phenotype of interest.
If you want to get a functional insight for an uncharacterized gene, you can use its network neighbors. YeastNet connects genes that are functionally associated. Therefore, the uncharacterized gene may have similar function of its connected network neighbors. This search page collects all known functional annotations (Gene Ontology biological process terms) among neighbors of a query gene on YeastNet, and then list the functional terms by the order of enrichment.
To identify modulators for a cell state (e.g., drug response), you may use differentially expressed genes (DEGs) upon transition of cell state (i.e., expression signature of the cell state). Many modulators, however, do not change their transcript level while responding cell state transition. To identify genetic modulators with no transcriptional change, we use genes highly connected to the DEGs for the cell state. This search page uses pre-defined gene sets composed of genes (central genes) and their network neighbors. Users need to submit DEGs for a specific cell state (e.g., genes responding to drug treatment). Then the search page conducts association analysis across all the pre-defined gene sets for a submitted set of DEGs. If a pre-defined gene set is significantly associated with the given DEGs, the central gene is a candidate modulator for the query cell state.>
(i) Find new members of a pathway
If you want to identify new candidate genes for a function/pathway/phenotype, you can start search by submitting a set of known genes for the function/pathway/phenotype. These submitted genes are used as 'seed genes' for network search. This search page collects closely connected genes to the seed genes on YeastNet by 'guilt-by-association' method and list the prioritized genes by total edge weight score (sum of log likelihood score) to the seed gene set. Highly ranked genes are good candidates for new members of a function/pathway/phenotype of interest.
(ii) Infer functions from network neighbors
If you want to get a functional insight for an uncharacterized gene, you can use its network neighbors. YeastNet connects genes that are functionally associated. Therefore, the uncharacterized gene may have similar function of its connected network neighbors. This search page collects all known functional annotations (Gene Ontology biological process terms) among neighbors of a query gene on YeastNet, and then list the functional terms by the order of enrichment.
(iii) Find modulators for a cell state
To identify modulators for a cell state (e.g., drug response), you may use differentially expressed genes (DEGs) upon transition of cell state (i.e., expression signature of the cell state). Many modulators, however, do not change their transcript level while responding cell state transition. To identify genetic modulators with no transcriptional change, we use genes highly connected to the DEGs for the cell state. This search page uses pre-defined gene sets composed of genes (central genes) and their network neighbors. Users need to submit DEGs for a specific cell state (e.g., genes responding to drug treatment). Then the search page conducts association analysis across all the pre-defined gene sets for a submitted set of DEGs. If a pre-defined gene set is significantly associated with the given DEGs, the central gene is a candidate modulator for the query cell state.>

| Back to Main |
